|
Link page added
A new Link Page has been created as promised
a while ago. It is still in its infancy but will hopefully
grow in the future. Contributions are encouraged.
It can be accessed from a new "Link" button on the left side of the
NMR home page.
Aside from some international links, there is a link to our own Mass Spec Lab as well as the X-Ray Laboratory.
Please keep in mind that spectrometer host computers do not
allow web browsing outside the U of A domain.
Solvents and
techniques update (GDQF- and GTQF-COSY)
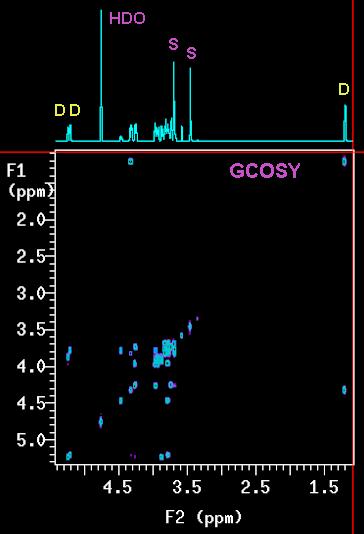
The gradient-enhanced COSY experiment (GCOSY)
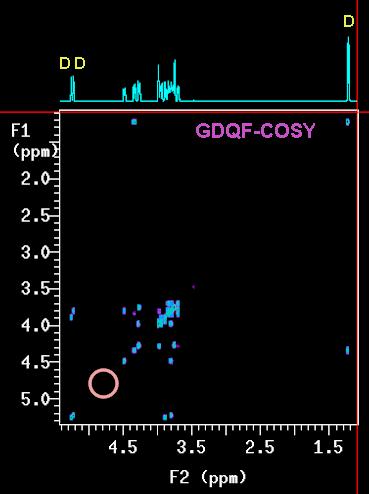
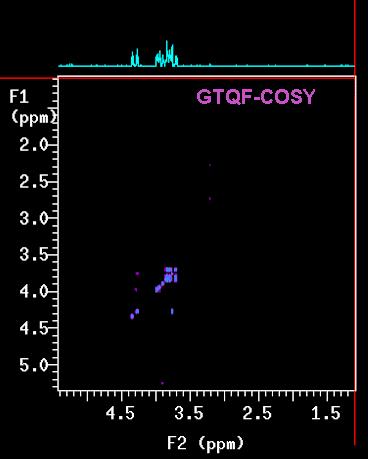
is a favourite to most users. Two variations of this technique, namely the double-quantum
filtered (GDQF-COSY) and triple-quantum filtered (GTQF-COSY) version
have been added. The techniques are, fortunately,
much easier to use
than their acronyms suggest.
As the names imply, some filtering is going on. i.e. you will
see less signals than with a simple GCOSY experiment. How much less is described here and
supplemented by examples,
a
summary Table and the pulse sequence is depicted as well.
First, to use these techniques use the button
GMQF-COSY in the EZ NMR S+A panel (M in GMQF is
used to represent the both the D and T version in one single name: multiple). The macro will calculate the required number of
transients based on your input for molecular weight and amount of sample. This will be
more than for the standard GCOSY, especially if you decide to run the triple-quantum
version by changing the quantum level parameter qlvl from its default value of 2 to
3.
You should also remember that the signals build up like a sine function
in F1, not
cosine like most NMR techniques. This means that if you process the data very early,
while the acquisition is still continuing, you may see very little signal or nothing at
all. For the same reason, linear prediction in F1 fails when used
too early in the acquisition
process: there is simply not enough information available to base the prediction on.
A
little bit of patience is a necessity for these techniques.
By far the most useful of the two is the
double quantum filtered version. It filters out all signals that are not coupled to anything else, in
other words singlets. First and foremost this is the
HDO residual signal but CDCl3 works as
well. In fact, the removal of the HDO signal is so efficient that a signal hidden
underneath the solvent peak, as long as it is not a singlet itself, will
(magically!) appear. This is
also useful when there are lots of signals from uncoupled methyl groups. These very
intense peaks often create undesired t1-noise that can interfere with the data analysis.
The GDQF-COSY will eliminate them all at once.
The next higher level (GTQF-COSY) removes everything the DQ version removes plus
also
doublets, or in general, signals that have only one coupling partner. What is really left
in the spectrum is somewhat complicated: some proton signals stay as diagonal peaks but
with no more correlation peaks (cross-peaks), whereas others survive the filtration
process and show normal diagonal and cross peaks. The on-line version of the Newsletter
contains a Table where this is summarized.
2D
processing: when to use what command
The processing (i.e. Fourier transformation) of 2D data
sets can be as simple as the command wft2d but, occasionally, depending on the
techniques used, it is much more complicated than that. For example, wft2d(1,0,-1,0,0,-1,0,-1)
is one command most people don't want to type in... You don't have to.
The WFT2D button in EZ NMR is "smart" enough to choose
the right format for the processing depending on what technique is in use. If you need to transform data only half way (along F2 but not F1), then use
the WFT2D F2 ONLY button.
This macro also knows what to use
as the proper coefficients for the spectrum at hand.
|