4. Sample preparation and pre-acquisition activities
4.1 Sample preparation
Filtering a sample prior to acquisition can greatly improve the quality of NMR spectra by removing contaminants that will adversely affect the spectral quality.
Contaminants that affect spectral quality in descending order of severity:
problem |
what to do |
paramagnetic substances, e.g. Ni, Mn |
filtration through Chelex column (Sep-Pak for carbohydrates *) |
solid or foreign material, e.g. septa pieces |
filtration through cotton wool or 0.22 mm microfilter |
high salt content |
filtration through Sep-Pak for carbohydrates * |
precipitate of dissolved compound |
filtration through cotton wool or microfilter; change solvent |
|
Filtration through cotton wool
* For carbohydrate Sep-Pak filtration see the Appendix at the end of this document Figure 4.2 : Filtration through cotton wool |
4.2 Sample volume and sample concentration
Correct sample volume is critical for good results.
- Too low sample volume can make it impossible to obtain high quality spectra; the sample can be impossible to shim; poor magnetic field homogeneity
- The time required for shimming increases dramatically or may fail with incorrect sample volume (especially when sample volume is too low)
- Too high sample volume unnecessarily dilutes the sample and may consume potentially costly deuterated solvents (i.e. THF-d8 ~$80/5g)
|
|
||
|
Agilent probes are 0.7 mL probes, hence the concentration of the sample in mM is given by:
conc [mM] = (mgx1000)/(m.w.x0.7)
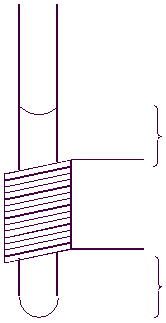
Ideal sample height is indicated by the
Figure 4.3 : NMR tube and sample volume |
||
4.3 Spectral References
Parameter sets use automatic referencing as shown below (strictly speaking, only valid at 27.0°C):
solvent |
H1 (number of lines) |
C13 (number of lines) |
CDCl3 |
7.26 ppm (1) |
77.0 ppm (3) |
C6D6 |
7.15 ppm (1) |
128.0 ppm (3) |
CD2Cl2 |
5.32 ppm (3) |
53.8 ppm (5) |
CD3OD |
3.30 ppm (5) |
49.0 ppm (7) |
DMSO |
2.49 ppm (5) |
39.5 ppm (7) |
D2O |
0.1% ext. acetone @ 2.225 ppm (1) |
1% ext. acetone @ 31.07 ppm (1) |
Ac-D6 |
2.04 ppm (5) |
29.8 ppm (7) |
H1 and C13 spectra of all 7 solvents (incl. common impurities) can be found at http://nmr.chem.ualberta.ca ® FAQ ® solvents: spectra of supported solvents.
Referencing to external acetone puts HDO at 4.75 ppm (27.0°C). The HDO chemical shift changes +/- 0.008 ppm for every -/+ 1.0°C (note +/- signs!).
If changes are made by the user, correct referencing can be restored by using the EZ NMR P+P panel Referencing: Adjust to Default whereby the referencing is restored in all supported solvents and experiments, 1D or 2D, homo- or heteronuclear.
Alternatively, if different referencing is desired and in unsupported solvents, the EZ NMR P+P panel Referencing: Adj to Line @ Cursor allows placement of the cursor on a spectral line and entering the desired value in ppm (1D and 2D).
4.4 Access to spectrometers, online reservations, etc.
v700: Robotic Sample Handling
i600: priority access to Carbohydrate Center and the research group of Dr. Vederas
i400, ibd5, u500: General access via self-registration and on-line reservation system at http://nmr.chem.ualberta.ca which enforces the reservation rules. Reservation rules are spectrometer specific but do change periodically, see nmr.chem.ualberta.ca for instrument specific details. The reservation rules for the ibd5 are given below as an example:
ibd5 reservation rules, every day (365 days a year):
- time slots are 15 minutes from 08:00 (8 a.m.) to 20:00 (8 p.m.)
- no booking restrictions from 20:00 (8 p.m.) until the next morning 08:00 (8 a.m.) time slot
- no reservations are allowed more than 24 hours in advance with the exception of tomorrow's overnight period which can be booked after 16:00 (4 p.m.)
- the 5 minute rule: if the spectrometer is booked but not used for more than 5 minutes of the starting time, the reservation is forfeit
- Monday - Friday: no more than four consecutive time slots per day between 08:00 (8 a.m.) and 20:00 (8 p.m.)
- Saturday and Sunday: up to twelve consecutive time slots (3 hours) but only once a day for any user between 08:00 (8 a.m.) and 20:00 (8 p.m.)
- Holidays and U of A closure days:reservation rules as Saturday and Sunday
Time pooling or combining your NMR time with another user is strictly prohibited and will result in withdrawal of your access to the NMR spectrometers.
4.5 Logging into the system and starting VNMRJ
Linux computers are multi-user machines that protect data for each user with a password.
It is critical that the correct user account is used for the proper function of the system and for saving and locating data.
Logging on to the system requires a username and password, these are group specific and case sensitive.
|
Figure 4.4: Linux login screen, enter username. |
|
Figure 4.5: Linux login screen, enter password. |
- a search of immense proportions for the data can result, especially if you don’t remember the exact filename, requiring orders of magnitude more time than a quick log out/log in
- the ownership of the file(s), if ever found, is wrong and requires intervention by the system administrator to allow you to do anything with the file(s)
- some macros act differently depending on the username
Using the correct username prevents writing the data in the wrong place and automatically provides the correct ownership of the data.
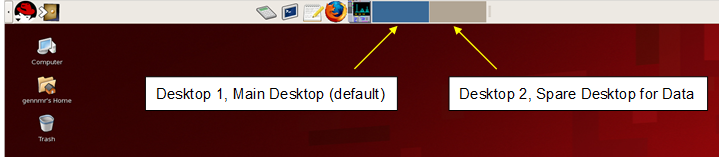
After logging in, one arrives at the Linux desktop, shown below. In fact, there are two desktops available to use, both are identical but it is recommended that the first desk top, or default, is used for VNMRJ and the second for data manipulations.
|
Figure 4.6 : Linux main (default) desktop |
4.6 The VNMRJ interface
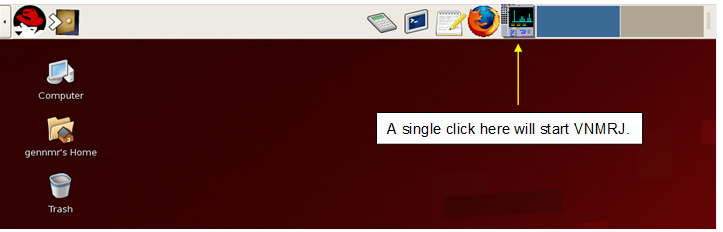
Start VNMRJ with a single click on the icon indicated below (DO NOT DOUBLE CLICK!):
 |
|
Figure 4.7 : Starting VNMRJ |
|
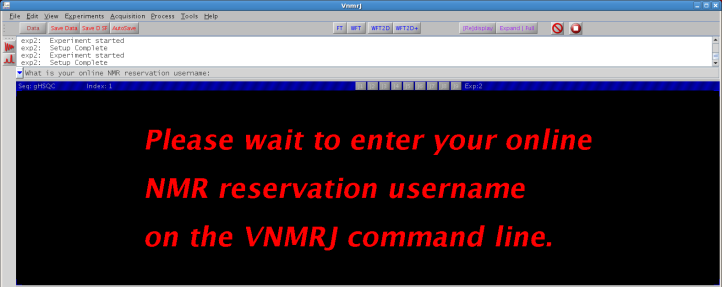
Research accounts require a valid online user reservation name to use VNMRJ:
|
Figure 4.8 : User identification |
Invalid input will close VNMRJ, a valid online reservation username provides the following information and makes the program available to use:
|
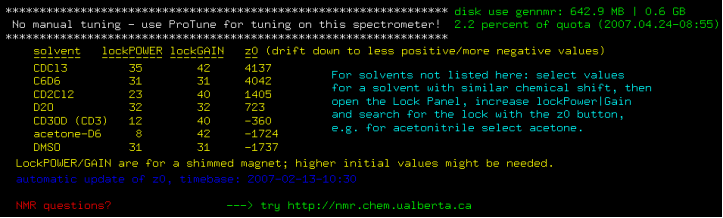
Figure 4.9 : z0 and disk usage information |
Tuning information, white lettering shown at the top of the message above, is spectrometer dependent, e.g. no tuning on m400, ProTune on i400, ibd5, u500, i600 and v700, manual tuning on i300.
Disk usage and disk quota information on the central data server is shown in green lettering top right hand corner of the message above. Surpassing the disk quota will prevent saving of data.
LockPower, lockGain and z0 information are spectrometer dependent! z0 slowly changes with time (magnet drift). The z0 information can be used as a guide when automated locking fails.
After starting VNMRJ, and identifying yourself to the system, you should arrive in the last experiment number used before exiting VNMRJ. VNMRJ has 9 experiments by default (additional experiments can be created by the user). The dark blue box indicates the currently joined experiment.
Use any of the grey buttons to join one of the 9 default experiments. |
|
Figure 4.10 : Join experiment buttons and currently joined experiment |
|
The VNMRJ interface is shown below.
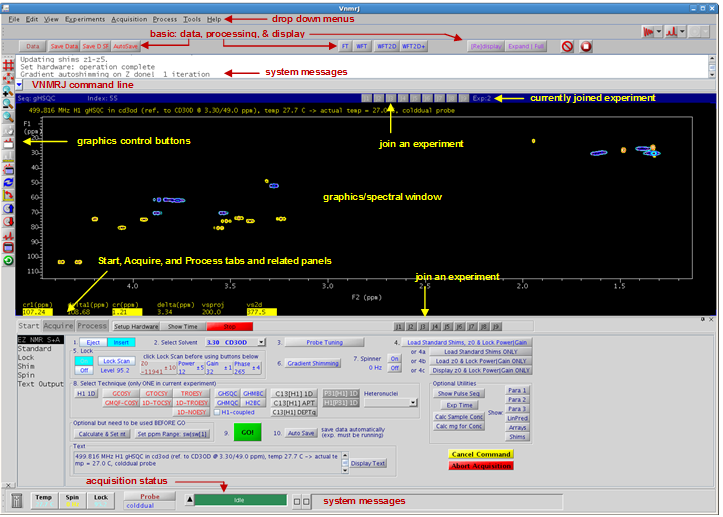 |
Figure 4.11 : The VNMRJ interface |
There are two fundamentally different ways to operate VNMRJ:
- changing parameters, entering commands and macros on the VNMRJ command line with the keyboard
- using buttons on panels with the mouse thereby choosing predefined parameter sets and executing predefined macros
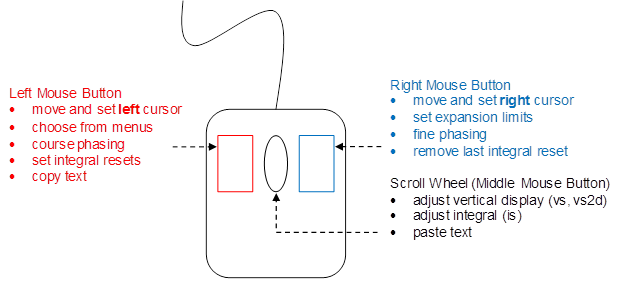
All Linux mice are 3-button button mice, the scroll wheel that acts as a middle button. The Linux mouse:
|
Figure 4.12: The Linux Mouse |
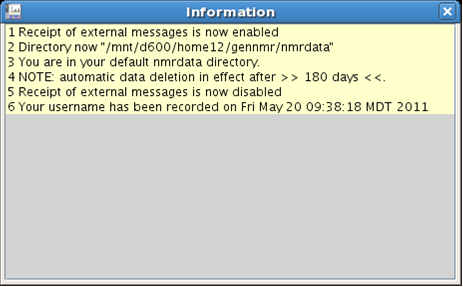
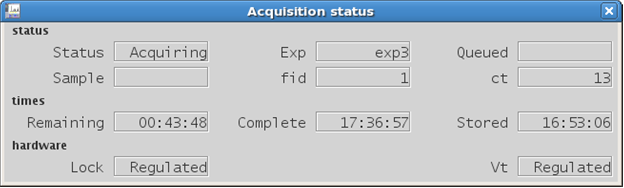
Messages coming from VNMRJ and the console (system messages) are limited to one to three lines. The acquisition status, outlined in Fig. 4.11, does not indicate which experiment is being acquired. Click on the buttons adjacent to these information boxes to open information and acquisition status windows. Place these windows on the second monitor and enjoy feedback from the spectrometer.
|
Figure 4.13 : The Information window |
|
Figure 4.14 : The Acquisition status window |